RNASeq Workflow Module
Material by Rajiv Mayani, Karan Vahi, Jennifer Herstein, Andrew Clark
RseqFlow
RseqFlow is an RNASeq analysis pipeline which offers an express implementation of analysis steps. It can perform pre and post mapping quality control (QC) for sequencing data, calculate expression levels for uniquely mapped reads, identify differentially expressed genes, and convert file formats for ease of visualization.
Introduction
Use a browser to open the tutorial on github, located at: http://github.com/arokem/2013-09-16-ISI/lessons/rnaseq/tutorial.html
A presentation on RNASeq that gives a high level overview of the process can be found here
A workflow is an automatic process, during which, data, information or tasks are passed from one module to another for action, according to a set of procedural rules. So with workflow, users can acquire analysis results expressly saving from running the pipeline step by step, inputting commands and arguments for each module. As pipeline builders, we can model, design, execute, debug, re-configure and re-run the analysis and visualization pipelines with workflow. For users, the workflow can not only execute a series of computational or data manipulation steps according to presetting steps automatically, but also enables to track the provenance of the workflow execution results, such as, methods used, machine calibrations and parameters, services and databases accessed, data sets used, etc.
The Pegasus Workflow Management Service was adopted to manage the workflow's operations. It helps workflow execute in different kinds of different environments including desktops, campus clusters, grids, and clouds. The Pegasus RseqFlow pipeline was deployed in a Virtual Machine, which exempts users from complex installation and configuration. If users want to deploy on their own cluster, they can use the VM as a submit machine to submit jobs into the cluster.
This tutorial walks through the steps of creating and running the Expression Estimation pipeline through Pegasus. The Virtual Machine has an input dataset for the tutorial workflow and all the executables preinstalled.
The RseqFlow Expression Estimation (EE) pipeline implements expression estimation at gene, exonic, and splice junction levels based on the alignment of reads to the transcriptome. The user supplies the pipeline with a gene annotation file, a FASTA reference file of transcriptome sequences and a FASTQ RNASeq dataset.
The EE pipeline aligns the FASTQ dataset to the transcriptome and keeps uniquely mapped reads for downstream analysis, i.e. those that align only to a single position in the transcriptome reference. Multi-mapped reads, those that align to multiple positions, are eliminated. The annotation file and the alignment output files are split into separate per chromosome files for faster parallel processing at the end of which the results are then merged back into a single file. Expression levels of genes, exons and splice junctions are calculated based on the number of unique reads aligned. For genes and exons, RPKM (Number of reads Per Kilobases per Million mapped reads) is calculated, and for splice junctions, RPM (Number of reads Per Million mapped reads) is calculated. The final output consists of three expression files, one each for genes, exons, and splice junctions.
The schematic of the EE pipeline is shown below
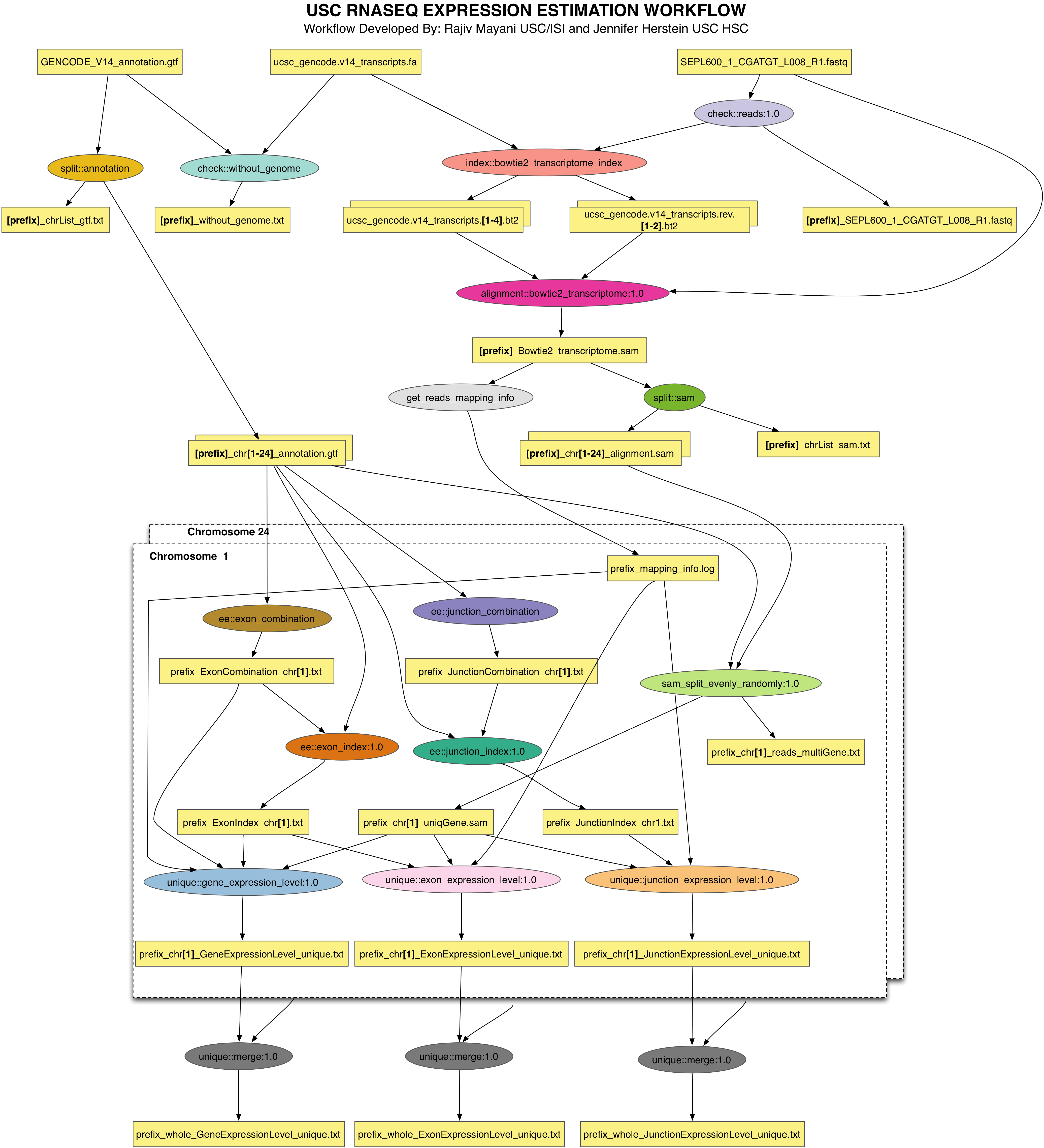
All of the steps in this tutorial are performed on the command-line. The convention we will use for command-line input and output is to put things that you should type in bold, monospace font, and to put the output you should get in a normal weight, monospace font, like this:
[user@host dir]$ you type this
you get this
Where [user@host dir]$ is the terminal prompt, the text you should type is "you type this", and the output you should get is "you get this". The terminal prompt will be abbreviated as $. Because some of the outputs are long, we don't always include everything. Where the output is truncated we will add an ellipsis '...' to indicate the omitted output.
If you are having trouble with this tutorial, or anything else related to RseqFlow , you can contact the USC RseqFlow mailing list at RseqFlow@isi.edu to get help.
Getting started
In order to reduce the amount of work required to get started we have provided several virtual machines that contain all of the software required for this tutorial. Virtual machine images are provided for VirtualBox. Information about starting the bootcamp VM is in the Virtualbox Lesson. Please follow the instructions for starting the VM before continuing with this tutorial.
Generating the workflow
In this tutorial, we will generate an EE workflow that runs across chromosomes 1-22, X, Y and M. The schema of the workflow that is generated is the same as shown above. It takes as input
- a FASTQ file of RNAseq reads
- 2 reference files, a FASTA file of transcript sequences and a GTF annotation file.
In the VM, the input data (451 MB) for the tutorial is present here
[tutorial@localhost data]$ cd /rnaseq/ee/data
[tutorial@localhost data]$ ls
GENCODE_V14_annotation.gtf SEP034_AAGGGA_L002_R1-75Kreads.fastq ucsc_gencode.v14_transcripts.fa
The workflow takes about 20-30 minutes to run in this VM.
In order to run the RseqFlow EE pipeline on this input dataset we need to generate a description of the workflow that can be executed through the workflow system. We will run the RseqFlow workflow generator expression_estimation.py to generate this description.
The VM has a helper generate_dax.sh script that passes the right arguments to the workflow generator for the canned example.
[tutorial@localhost data]$ cd /rnaseq/ee/
[tutorial@localhost ee]$ ./generate_dax.sh rnaseq.dax
Generating Pegasus DAX
+ expression_estimation.py --output-prefix prefix --unique --fastq SEP034_AAGGGA_L002_R1-75Kreads.fastq --transcriptome ucsc_gencode.v14_transcripts.fa --annotation GENCODE_V14_annotation.gtf --bin-dir bin --dax rnaseq.dax --verbose
2013-09-12 20:04:10,237 - Chromosomes included X, Y, M, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22
2013-09-12 20:04:10,237 - Add jobs to check input files
2013-09-12 20:04:10,238 - Add jobs to split files by chromosomes
2013-09-12 20:04:10,239 - Add jobs to build exon index and combination files
2013-09-12 20:04:10,243 - Add jobs to build junction index and combination files
2013-09-12 20:04:10,249 - Add jobs to compute unique expression level
In the above example, the input files to use are specified by passing the --fastq, --annotation and --transcriptome options. You just need to specify the basename of the input files. The --bin-dir option specifies where all the executables required for this workflow are installed.
The executables referred to by this workflow are in the bin directory.
[tutorial@localhost ee]$ ls /rnaseq/ee/bin
bowtie2-align GeneExpressionLevel.py
SplitByChromosome_for_transcriptomeSamFile.py
GeneExpressionLevelmappable.py
SplitByChromosome_for_transcriptomeSequenceFaFile.py
GetReadsFromTranscripts.py
bowtie2-build JunctionCombination.py
Merge.sh
GeneExpressionLevel_proportionAssign.py
Check_for_reference_annotation_withoutGenome.py
ModifyExonIndex.py
....
Pegasus reads workflow descriptions from DAX files. The term "DAX" is short for "Directed Acyclic Graph in XML". DAX is an XML file format that has syntax for expressing jobs, arguments, files, and dependencies. Let's look at the generated DAX for the workflow
more rnaseq.dax
...
The generated DAX has three main sections
The locations of the executables that are referred to by the workflow. These can be in a separate tranformation catalog file also.
The jobs making up the workflow. Notice, how jobs only refer to the input and output files by logical identifiers.
The edges between the jobs in the workflow that specify the underlying DAG ( Directed Acyclic Graph).
Note how the DAX is devoid of any data movement jobs. These are added by Pegasus when the DAX is planned to an executable workflow.
Running the workflow in the VM
The planning stage is where Pegasus maps the abstract DAX to one or more execution sites.
The pegasus-plan command is used to plan a workflow. This command takes quite a few arguments, so we created a run_dax.sh wrapper script that has all of the arguments required for the RseqFlow EE workflow:
[tutorial@localhost ee]$ ./run_dax.sh rnaseq.dax
Planning Pegasus workflow
+ pegasus-plan -Dpegasus.catalog.replica.directory.site=condor_pool --dax rnaseq.dax --dir submit --conf properties --sites condor_pool --nocleanup --cluster horizontal --output-site local --input-dir data --submit
2013.09.12 21:06:13.756 EDT: Submitting job(s).
2013.09.12 21:06:13.795 EDT: 1 job(s) submitted to cluster 156.
2013.09.12 21:06:13.853 EDT:
2013.09.12 21:06:13.865 EDT: -----------------------------------------------------------------------
2013.09.12 21:06:13.875 EDT: File for submitting this DAG to Condor : expression_estimation_prefix-0.dag.condor.sub
2013.09.12 21:06:13.885 EDT: Log of DAGMan debugging messages : expression_estimation_prefix-0.dag.dagman.out
2013.09.12 21:06:13.895 EDT: Log of Condor library output : expression_estimation_prefix-0.dag.lib.out
2013.09.12 21:06:13.905 EDT: Log of Condor library error messages : expression_estimation_prefix-0.dag.lib.err
2013.09.12 21:06:13.912 EDT: Log of the life of condor_dagman itself : expression_estimation_prefix-0.dag.dagman.log
2013.09.12 21:06:13.918 EDT:
2013.09.12 21:06:13.925 EDT: -----------------------------------------------------------------------
2013.09.12 21:06:13.939 EDT:
2013.09.12 21:06:13.949 EDT: Your workflow has been started and is running in the base directory:
2013.09.12 21:06:13.958 EDT:
2013.09.12 21:06:13.965 EDT: /rnaseq/ee/submit/tutorial/pegasus/expression_estimation_prefix/run0002
2013.09.12 21:06:13.981 EDT:
2013.09.12 21:06:13.995 EDT: *** To monitor the workflow you can run ***
2013.09.12 21:06:14.001 EDT:
2013.09.12 21:06:14.010 EDT: pegasus-status -l /rnaseq/ee/submit/tutorial/pegasus/expression_estimation_prefix/run0002
2013.09.12 21:06:14.016 EDT:
2013.09.12 21:06:14.021 EDT: *** To remove your workflow run ***
2013.09.12 21:06:14.053 EDT:
2013.09.12 21:06:14.059 EDT: pegasus-remove /rnaseq/ee/submit/tutorial/pegasus/expression_estimation_prefix/run0002
2013.09.12 21:06:14.069 EDT:
2013.09.12 21:06:15.369 EDT: Time taken to execute is 5.26 seconds
The script invokes the pegasus-plan command with arguments for the configuration file (--conf), the DAX file (-d), the submit directory (--dir), the execution site (--sites), the input directory where the input files are (--input-dir), the output site (-o) and (--submit) to submit the workflow for execution. For this workflow, (--cluster) option is also enabled that allows us to cluster the short running jobs (unique::exon_expression_level, unique::junction_expression_level, unique::gene_expression_level) together.
Memory intensive jobs
For the purposes of this workshop, the basecase for the VM is to run the tutorial in 512MB RAM. The workflow has the bowtie transcriptome index job that requires about 4GB of RAM to create an index from the reference files.
Hence, for this tutorial the index files are alredy present in the input directory.
[tutorial@localhost ee]$ ls -lht /rnaseq/ee/data/
total 731M
-rw-rw-r-- 1 tutorial tutorial 75M Sep 12 22:44 ucsc_gencode.v14_transcripts.rev.1.bt2
-rw-rw-r-- 1 tutorial tutorial 44M Sep 12 22:44 ucsc_gencode.v14_transcripts.rev.2.bt2
-rw-rw-r-- 1 tutorial tutorial 75M Sep 12 22:33 ucsc_gencode.v14_transcripts.1.bt2
-rw-rw-r-- 1 tutorial tutorial 44M Sep 12 22:33 ucsc_gencode.v14_transcripts.2.bt2
-rw-rw-r-- 1 tutorial tutorial 815K Sep 12 22:22 ucsc_gencode.v14_transcripts.3.bt2
-rw-rw-r-- 1 tutorial tutorial 44M Sep 12 22:22 ucsc_gencode.v14_transcripts.4.bt2
-rw-rw-r-- 1 tutorial tutorial 19M Sep 11 14:29 SEP034_AAGGGA_L002_R1-75Kreads.fastq
-rw-r--r-- 1 tutorial tutorial 243M Jun 12 16:47 GENCODE_V14_annotation.gtf
-rw-r--r-- 1 tutorial tutorial 190M Jun 12 16:47 ucsc_gencode.v14_transcripts.fa
The presence of the bt2 files enables Pegasus to skip the bowtie2transcriptomeindex_ID0000003 job as the outputs for it already exist.
When you ran run_dax.sh in the log you would have seen the following:
2013.09.13 00:15:36.802 EDT: [INFO] event.pegasus.reduce dax.id expression_estimation_prefix_0 - STARTED
2013.09.13 00:15:36.824 EDT: [INFO] Nodes/Jobs Deleted from the Workflow during reduction
2013.09.13 00:15:36.825 EDT: [INFO] bowtie2_transcriptome_index_ID0000003
2013.09.13 00:15:36.825 EDT: [INFO] Nodes/Jobs Deleted from the Workflow during reduction - DONE
Details about Pegasus Data Reuse can be found here.
If you want the index to be created from the reference files as part of the workflow simply remove the .bt2 files from the /rnaseq/ee/data/ directory.
Monitoring the Workflow
After the workflow has been submitted you can monitor it using the pegasus-status command:
[tutorial@localhost ee]$ pegasus-status submit/tutorial/pegasus/expression_estimation_prefix/run0001/
STAT IN_STATE JOB
Run 10:02 expression_estimation_prefix-0
Run 03:28 |--without_genome_ID0000001
Idle 03:11 |--stage_out_remote_condor_pool_0_0
Summary: 3 Condor jobs total (I:1 R:2)
UNREADY READY PRE QUEUED POST SUCCESS FAILURE %DONE
154 0 0 3 0 5 0 3.1
Summary: 1 DAG total (Running:1)
This command shows the workflow and the running jobs (in the above output it shows the expressionestimationprefix-0 job). It also gives statistics on the number of jobs in each state and the percentage of the jobs in the workflow that have finished successfully.
Use the watch option to continuously monitor the workflow:
[tutorial@localhost ee]$ pegasus-status -w submit/tutorial/pegasus/expression_estimation_prefix/run0001/
...
You should see all of the jobs in the workflow run one after the other. After about 20 minutes you will see:
(no matching jobs found in Condor Q)
UNREADY READY PRE QUEUED POST SUCCESS FAILURE %DONE
0 0 0 0 0 162 0 100.0
Summary: 1 DAG total (Success:1)
That means the workflow is finished successfully. You can type ctrl-c to terminate the watch command.
If the workflow finished successfully you should see the output files in the output directory. This file was created by the various transformations in the workflow and shows all of the executables that were invoked by the workflow:
[tutorial@localhost ee]$ ls -lht outputs/
-rw-r--r-- 1 tutorial tutorial 111 Sep 13 01:36 outputs/prefix_whole_JunctionExpressionLevel_unique.txt
-rw-r--r-- 1 tutorial tutorial 2.1M Sep 13 01:36 outputs/prefix_whole_ExonExpressionLevel_unique.txt
-rw-r--r-- 1 tutorial tutorial 278K Sep 13 01:36 outputs/prefix_whole_GeneExpressionLevel_unique.txt
-rw-r--r-- 1 tutorial tutorial 0 Sep 13 01:28 outputs/prefix_without_genome.txt
-rw-r--r-- 1 tutorial tutorial 29 Sep 13 01:27 outputs/prefix_SEP034_AAGGGA_L002_R1-75Kreads.fastq
Of these, the first three files (with prefix prefix\whole\) will be of interest to scientists.
Collecting Statistics
The pegasus-statistics command can be used to gather statistics about the runtime of the workflow and its jobs. The -s all argument tells the program to generate all statistics it knows how to calculate:
[tutorial@localhost ee]$ pegasus-statistics -s all submit/tutorial/pegasus/expression_estimation_prefix/run0001/
#
# Pegasus Workflow Management System - http://pegasus.isi.edu
#
# Workflow summary:
# Summary of the workflow execution. It shows total
# tasks/jobs/sub workflows run, how many succeeded/failed etc.
# In case of hierarchical workflow the calculation shows the
# statistics across all the sub workflows.It shows the following
# statistics about tasks, jobs and sub workflows.
# * Succeeded - total count of succeeded tasks/jobs/sub workflows.
# * Failed - total count of failed tasks/jobs/sub workflows.
# * Incomplete - total count of tasks/jobs/sub workflows that are
# not in succeeded or failed state. This includes all the jobs
# that are not submitted, submitted but not completed etc. This
# is calculated as difference between 'total' count and sum of
# 'succeeded' and 'failed' count.
# * Total - total count of tasks/jobs/sub workflows.
# * Retries - total retry count of tasks/jobs/sub workflows.
# * Total+Retries - total count of tasks/jobs/sub workflows executed
# during workflow run. This is the cumulative of retries,
# succeeded and failed count.
# Workflow wall time:
# The walltime from the start of the workflow execution to the end as
# reported by the DAGMAN.In case of rescue dag the value is the
# cumulative of all retries.
# Workflow cumulative job wall time:
# The sum of the walltime of all jobs as reported by kickstart.
# In case of job retries the value is the cumulative of all retries.
# For workflows having sub workflow jobs (i.e SUBDAG and SUBDAX jobs),
# the walltime value includes jobs from the sub workflows as well.
# Cumulative job walltime as seen from submit side:
# The sum of the walltime of all jobs as reported by DAGMan.
# This is similar to the regular cumulative job walltime, but includes
# job management overhead and delays. In case of job retries the value
# is the cumulative of all retries. For workflows having sub workflow
# jobs (i.e SUBDAG and SUBDAX jobs), the walltime value includes jobs
# from the sub workflows as well.
------------------------------------------------------------------------------
Type Succeeded Failed Incomplete Total Retries Total+Retries
Tasks 209 0 0 209 0 209
Jobs 161 0 0 161 0 161
Sub-Workflows 0 0 0 0 0 0
------------------------------------------------------------------------------
Workflow wall time : 11 mins, 18 secs
Workflow cumulative job wall time : 7 mins, 22 secs
Cumulative job walltime as seen from submit side : 8 mins, 43 secs
Summary : submit/tutorial/pegasus/expression_estimation_prefix/run0001/statistics/summary.txt
Workflow execution statistics : submit/tutorial/pegasus/expression_estimation_prefix/run0001/statistics/workflow.txt
Job instance statistics : submit/tutorial/pegasus/expression_estimation_prefix/run0001/statistics/jobs.txt
Transformation statistics : submit/tutorial/pegasus/expression_estimation_prefix/run0001/statistics/breakdown.txt
Time statistics : submit/tutorial/pegasus/expression_estimation_prefix/run0001/statistics/time.txt
Note: If the memory intensive job had been enabled, the walltime would be much longer.
Conclusion
Congratulations! You have completed the tutorial. For more information on the pipeline, please contact
Please contact the USC RseqFlow Mailing list at rseqflow@isi.edu if you need help.
