Non-linear Models
Learning Objectives
- Learners can identify a nonlinear model, and discuss the differences between linear and nonlinear models
- Learners can use
scipy.optimizeto fit a nonlinear model to data. - Learners can calculate and display model residuals for nonlinear models
Unfortunately, linear models don’t always fit our data. As shown before, they might produce large and systematic fit error, or they might produce parameter values that don’t make sense. In these cases, we might need to fit a non-linear model to our data. Non-linear models are mathematically expressed as:
\(\bf{y} = f(\bf{x}_1, \bf{x}_2, ... \bf{x}_n, \bf{\beta}) + \epsilon\),
Where \(f()\) can be almost any function you can think of that takes \(x\)’s and \(\beta\)’s as input. The problem is that, in contrast to the linear models, there is no formula that you can just plug your function and data into and derive the values of the parameters.
One way to find the value of the parameters for a given model is to use optimization. In this process, the computer systematically tries out different values of \(\beta\) and finds a set of these values that minimizes the SSE relative to the data.
Introducing scipy
The scipy library contains many functions that are useful in a variety of scientific computing tasks.
If you import the library, you will see that it is composed of a variety of sub-modules, each devoted to a different set of algorithms, or a topic:
SciPy: A scientific computing package for Python
================================================
Documentation is available in the docstrings and
online at http://docs.scipy.org.
Contents
--------
SciPy imports all the functions from the NumPy namespace, and in
addition provides:
Subpackages
-----------
Using any of these subpackages requires an explicit import. For example,
``import scipy.cluster``.
::
cluster --- Vector Quantization / Kmeans
fftpack --- Discrete Fourier Transform algorithms
integrate --- Integration routines
interpolate --- Interpolation Tools
io --- Data input and output
linalg --- Linear algebra routines
linalg.blas --- Wrappers to BLAS library
linalg.lapack --- Wrappers to LAPACK library
misc --- Various utilities that don't have
another home.
ndimage --- n-dimensional image package
odr --- Orthogonal Distance Regression
optimize --- Optimization Tools
signal --- Signal Processing Tools
sparse --- Sparse Matrices
sparse.linalg --- Sparse Linear Algebra
sparse.linalg.dsolve --- Linear Solvers
sparse.linalg.dsolve.umfpack --- :Interface to the UMFPACK library:
Conjugate Gradient Method (LOBPCG)
sparse.linalg.eigen --- Sparse Eigenvalue Solvers
sparse.linalg.eigen.lobpcg --- Locally Optimal Block Preconditioned
Conjugate Gradient Method (LOBPCG)
spatial --- Spatial data structures and algorithms
special --- Special functions
stats --- Statistical Functions
Utility tools
-------------
::
test --- Run scipy unittests
show_config --- Show scipy build configuration
show_numpy_config --- Show numpy build configuration
__version__ --- Scipy version string
__numpy_version__ --- Numpy version string
To use any one of these sub-modules, we’ll need to import it specifically.
For example:
This module contains many functions that deal with various optimization tasks and implement many different algorithms for optimization. We’ll focus here on one of these functions, curve_fit, which systemtatically searches for the parameters that mimize the squared errors of a function relative to data.
Optimization in practice
Let’s now consider the steps you will need to take in fitting a non-linear model.
Defining the model
To perform optimization, we need to define the functional form of our model. For this kind of data, a common model to use (e.g in work by Yu and Levi) is a cumulative Gaussian function. The Gaussian distribution is parameterized by 2 numbers, the mean and the variance, so as in the linear model shown above, this model has 2 parameters:
\(y(x) = \frac{1}{2}[1 + erf(\frac{x-\mu}{\sigma \sqrt{2} })]\),
where \(erf\) is the so-called ‘error function’, that is implemented in scipy.special (from scipy import special).
from scipy import special
def cumgauss(x, mu, sigma):
"""
The cumulative Gaussian at x, for the distribution with mean mu and
standard deviation sigma.
Parameters
----------
x : float or array
The values of x over which to evaluate the cumulative Gaussian function
mu : float
The mean parameter. Determines the x value at which the y value is 0.5
sigma : float
The variance parameter. Determines the slope of the curve at the point of
Deflection
Returns
-------
Notes
-----
Based on:
http://en.wikipedia.org/wiki/Normal_distribution#Cumulative_distribution_function
"""
return 0.5 * (1 + special.erf((x-mu)/(np.sqrt(2)*sigma)))The doc-string already tells you one reason that we might want to use this function for this kind of data: one of the parameters of the function (mu) is simply the definition of the PSE. So, if we find this parameter, we already have the PSE in hand, without having to do any extra algebra.
Let’s plot a few exemplars of this function, just to get a feel for them:
fig, ax = plt.subplots(1)
ax.plot(x, cumgauss(x, 0.5, 0.25), label='mu=0, sigma=0.25')
ax.plot(x, cumgauss(x, 0.5, 0.5), label='mu=0, sigma=0.5')
ax.plot(x, cumgauss(x, 0.5, 0.75), label='mu=0, sigma=0.75')
ax.plot(x, cumgauss(x, 0.3, 0.25), label='mu=0.3, sigma=0.25')
ax.plot(x, cumgauss(x, 0.7, 0.25), label='mu=0.3, sigma=0.25')
ax.set_ylim([-0.1, 1.1])
ax.set_xlim([-0.1, 1.1])
ax.grid('on')
fig.set_size_inches([8,8])
plt.legend(loc='lower right')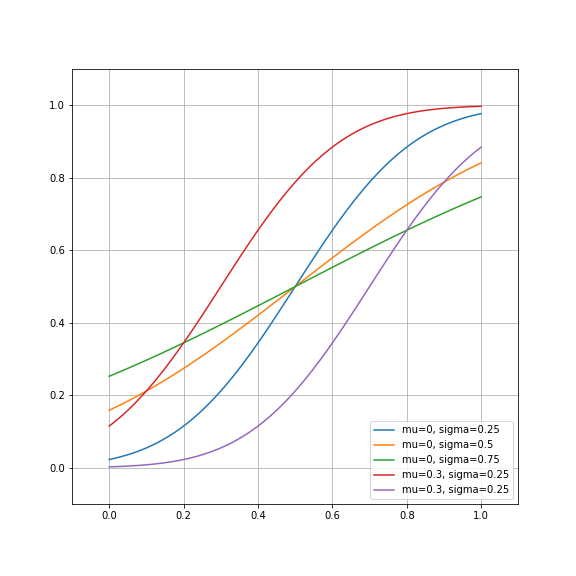
Optimizing and finding the parameters
To find the best parameters for the data, we will use the curve_fit function. This function takes as input a function, and data:
The first output contains the parameter estimates, and the second output is their covariance. This might be useful to know, but using the covariance is beyond the scope of this lesson.
As with the linear model, we would like to see how well the data fits the model.
Non-linear model fit
- Write the code that plots the model estimate and the actual data.
- Calculate the residuals and SSE of this model.
- What is the PSE of this model for both conditions (orthogonal and parallel)? Is this model better than the linear model?
In the next section we will see that there is more than even two ways to skin this cat. We’ll also introduce a powerful and general method for comparisons between different models, called cross-validation.