Data sharing dilemmas
October, 4th 2018
BRAIN Initiative Working Group 2.0 Workshop
Ariel Rokem, The University of Washington eScience Institute
Follow along at:



"How aggressive should we be about data sharing, task standardization and data aggregation?"
"Is data sharing working well enough? If not, why not?"
"How should NIH help to coordinate computational infrastructure?"
"How aggressive should we be about data sharing, task standardization and data aggregation?"
What is the value of making data and code available?
Reproducibility
"In god we trust, the rest bring data"- Deming
Reproducibility in the age of computational science
"An article about computational result is advertising, not scholarship. The actual scholarship is the full software environment, code and data that produced the result."
What is the value of reproducible research?
Reproducibility
Public access
Improved research quality
Greater impact
About 10% higher citation rate
Reuse, extension, interoperability
"How aggressive should we be about data sharing, task standardization and data aggregation?"
If the data was worth collecting, it is worth sharing
"Is data sharing working well enough? If not, why not?"
What would it take?
Standards
Software
Compute platforms
Socio-technical: training, incentives, careers
BIDS: a scientist-friendly data standard
The Brain Imaging Data Standard
Supported by INCF
Developed through an open and collaborative design process
An ongoing process
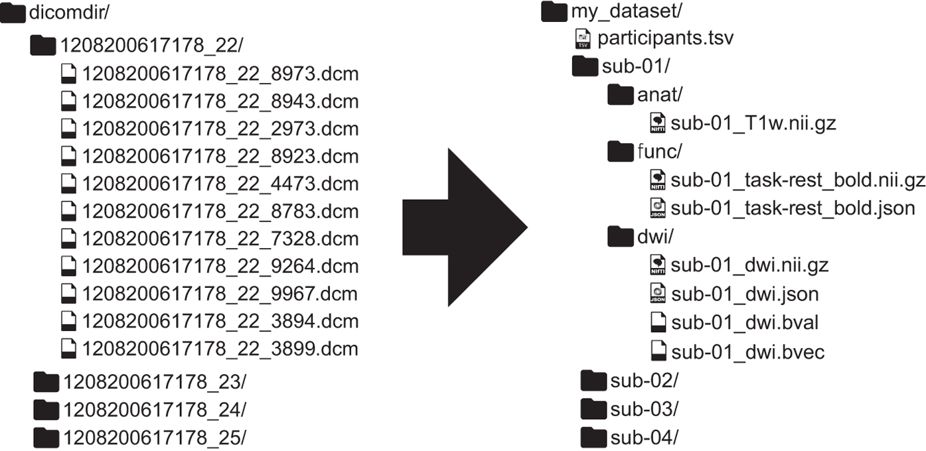
What does this enable?
Minimal curation => reusability
Validation: human and machine readable
Development of automated analysis tools
"How should NIH help to coordinate computational infrastructure?"
What are the requirements?
Bring the compute to the data
Scalable computing
Provide useful tools and interfaces
Facilitate interoperability (between datasets, between software libraries)
Control access
To the cloud!
Co-localization of data and compute
Scaling and elasticity
Consistent and open to all
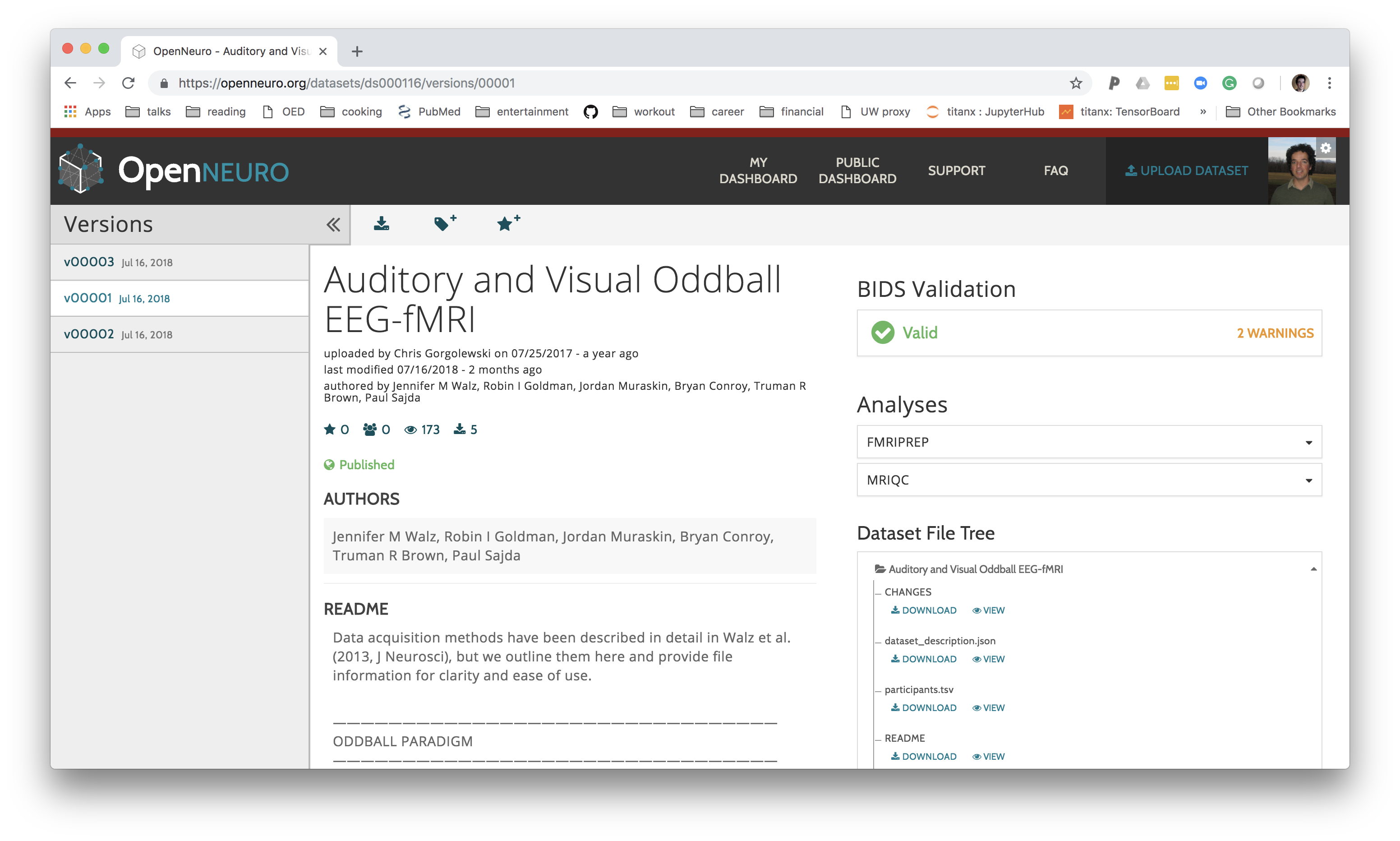
OpenNeuro
BrainLife
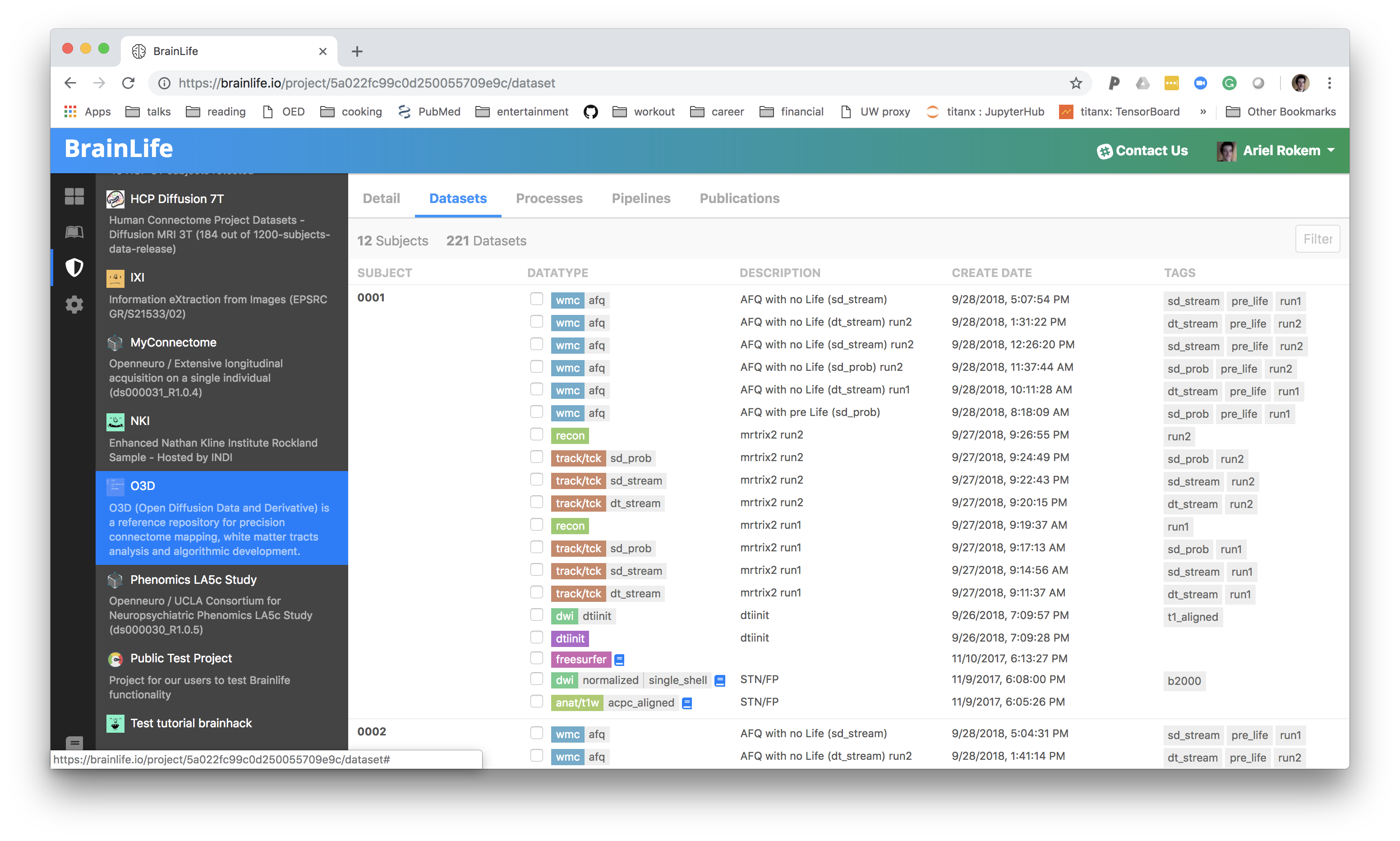
Do we need to build a portal for every collaboration in the BRAIN Initiative?
Open-source software for science
Python: an ecosystem for scientific computing
Free and open source
High-level interpreted language
Very wide adoption
Both in academic research and in industry
Python in Astronomy (ADS)
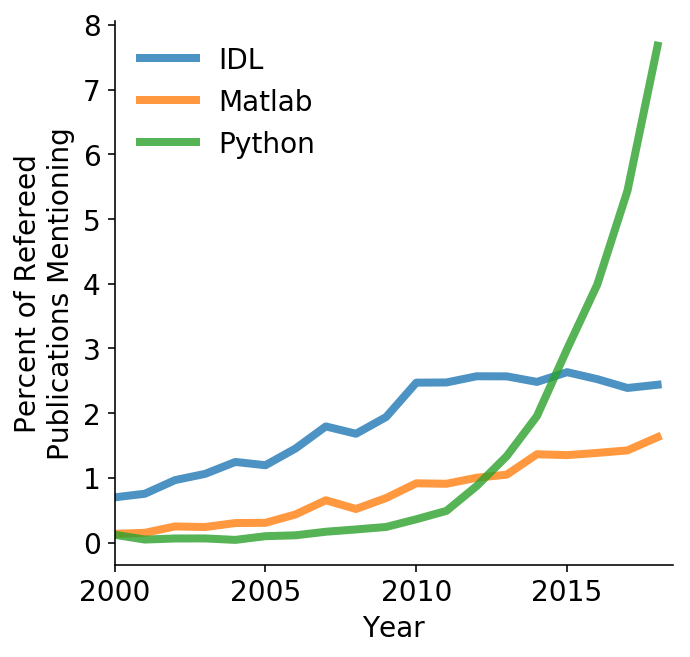
Grew out of IPython (an interactive Python shell)
Awarded the 2018 ACM Software System Award
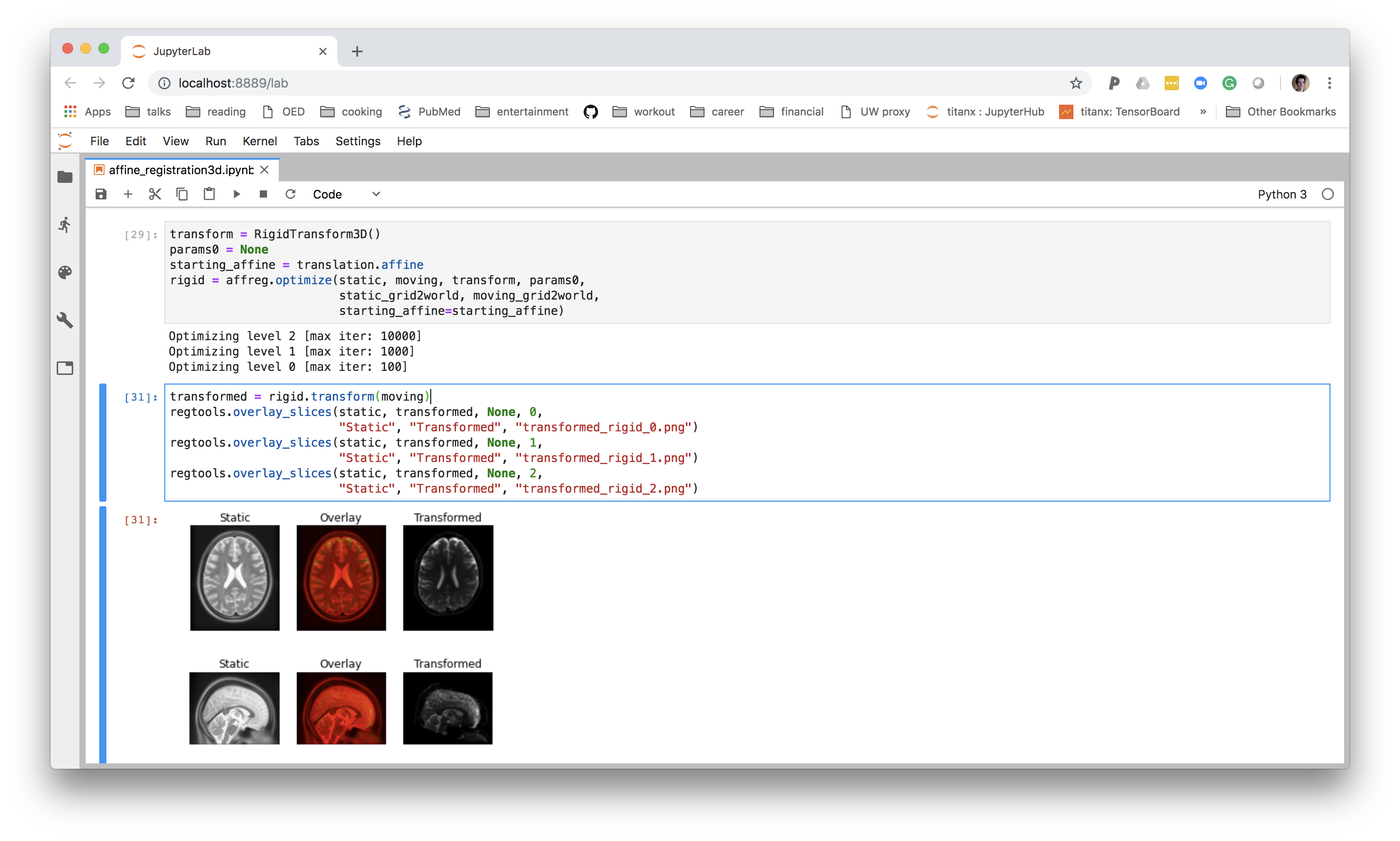
The Jupyter notebook
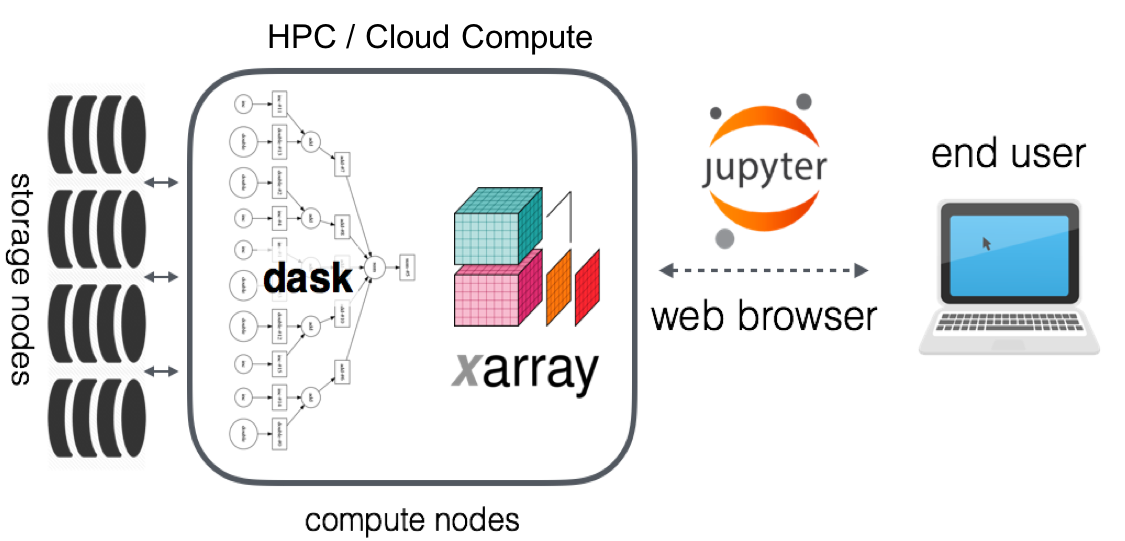
Cloud computing

Kubernetes
Vendor agnostic
Public cloud, HPC, combinations
Arguments against availability of data and code
Focusing on the wrong things
Waste of effort and resources
No immediate utility
Provides a disincentive to doing hard/risky experiments
Ways to mitigate these objections
Reasonable data embargos
Provide incentives for publication of valuable data and analysis
Make data and code publication easier
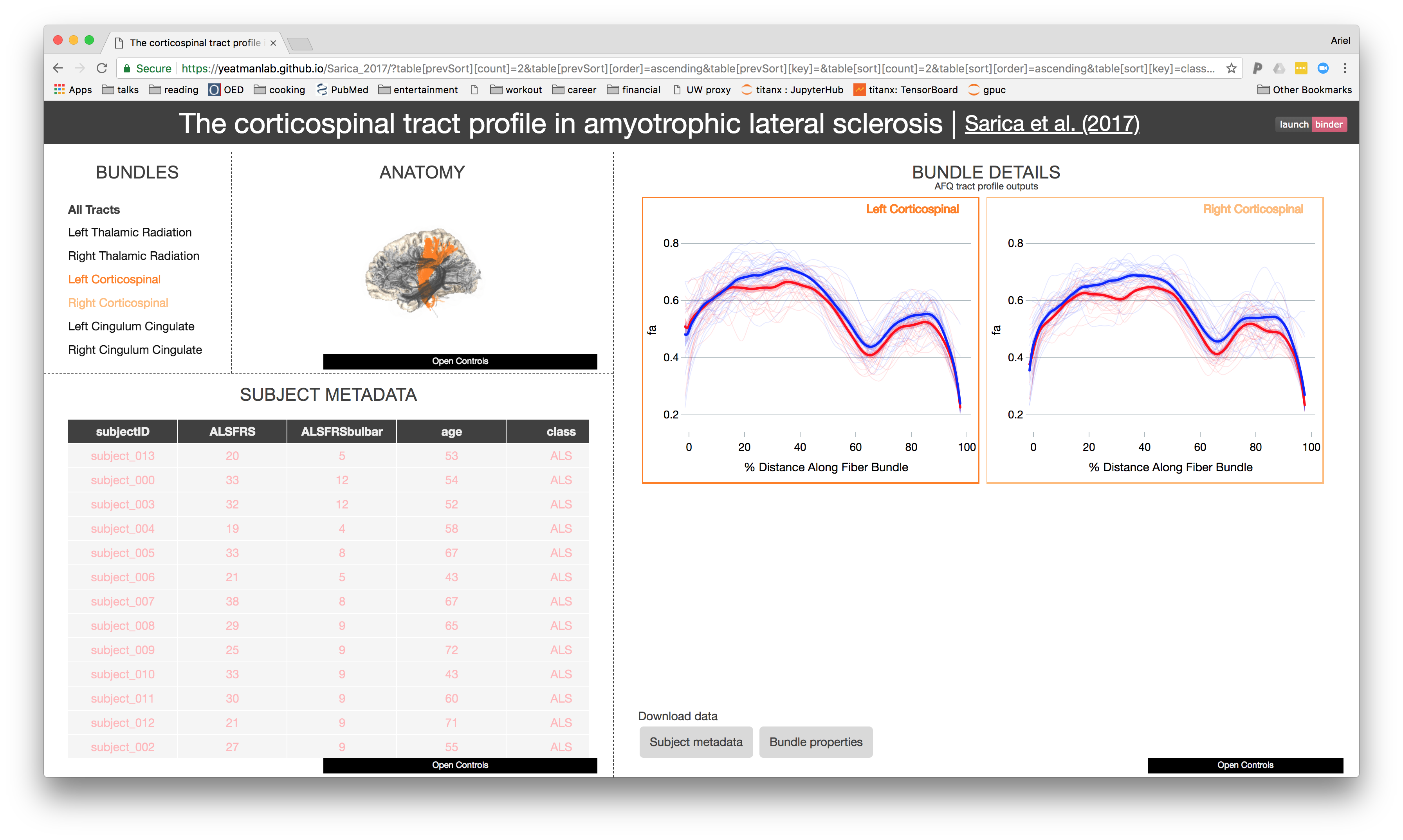
Publish useful extractions from the data
AFQ-Browser
Automatic data publication
Further exploration
Training for data science and neuroscience
Methods in data science are rapidly changing
Tools and practcies that are not usually part of the standard neuroscience curriculum
Learning often requires substantial hands-on experience
Communities of practice and training

"How aggressive should we be about data sharing, task standardization and data aggregation?"
"Is data sharing working well enough? If not, why not?"
"How should NIH help to coordinate computational infrastructure?"
Thanks!









Contact information




