PanNeuro: leveraging a community-based approach for big data neuroscience
BRAIN Initiative PI meeting, April, 2019
Ariel Rokem
The University of Washington eScience Institute
Follow along at:



Computational and circuit mechanisms underlying rapid learning
NINDS/NIH U19 funded through the BRAIN Initiative


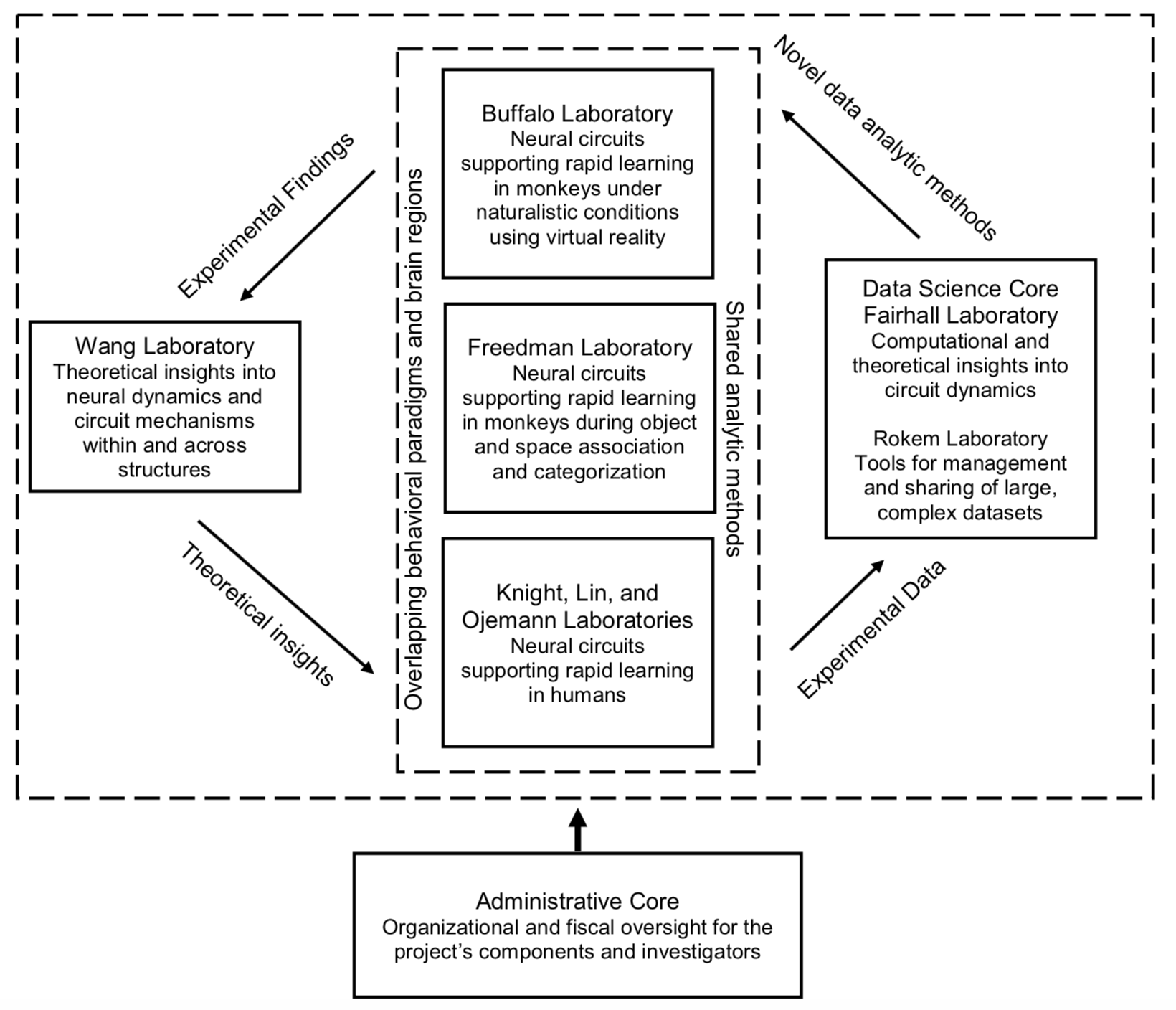
Aims
Identify the neural mechanisms that support schema development and rapid learning in association and categorization paradigms in monkeys and humans.
Develop and validate novel techniques for large-scale single unit recordings from multiple distributed regions of the nonhuman and human primate brain, during learning, through reversible inactivation, and during sleep.
Generate and test a multi-region computational understanding of circuit mechanisms that underlie schema development and rapid learning.
Learning to learn
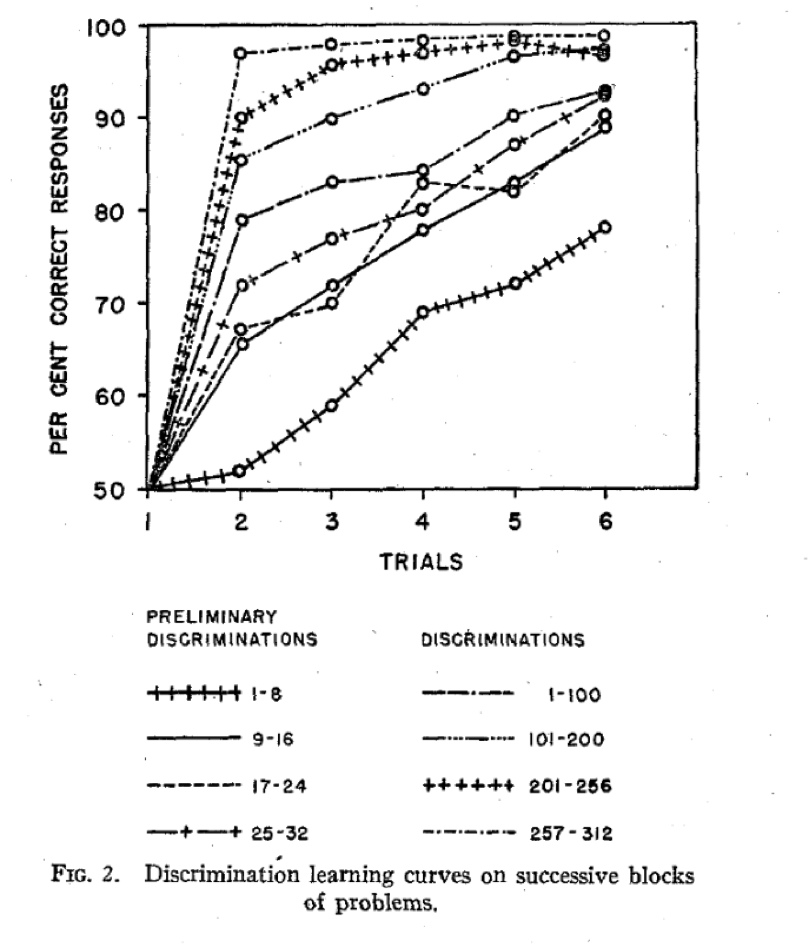
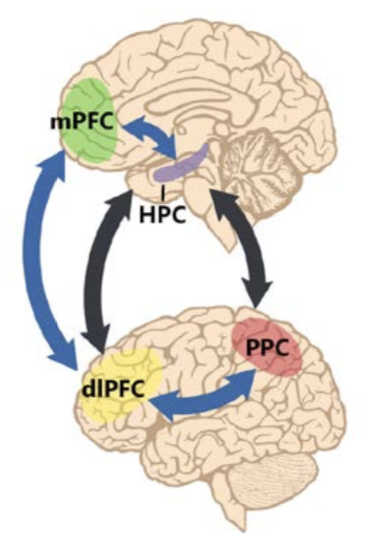
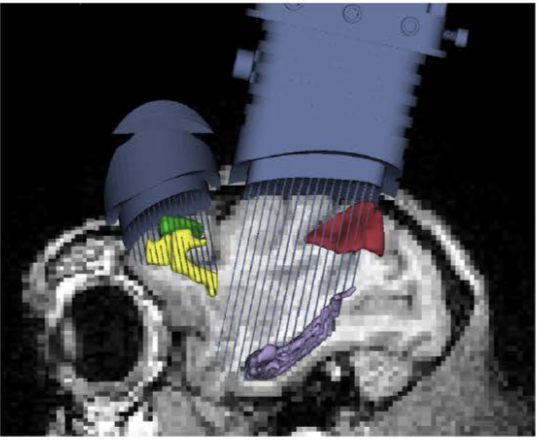
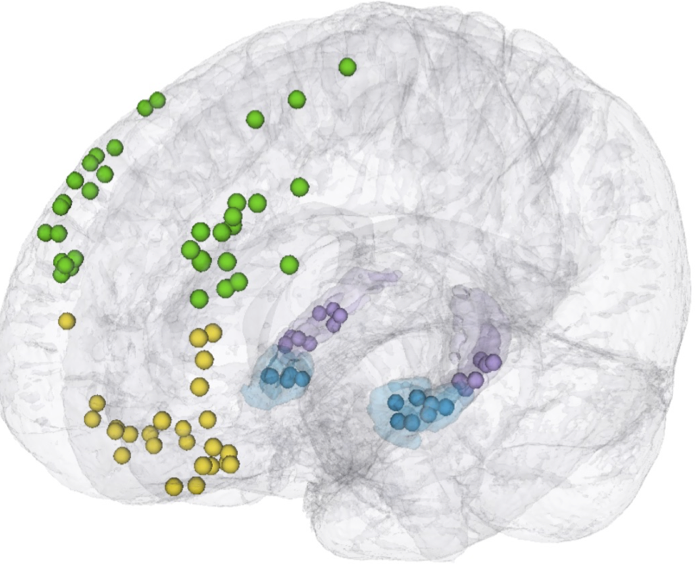
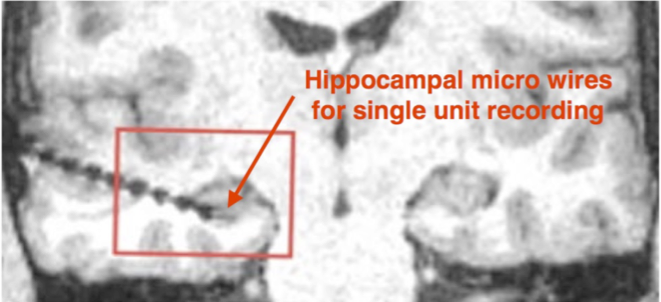


Data types:
Behavioral results
Non-human primate multichannel recordings
Human grid and electrode recordings
Model and simulation results
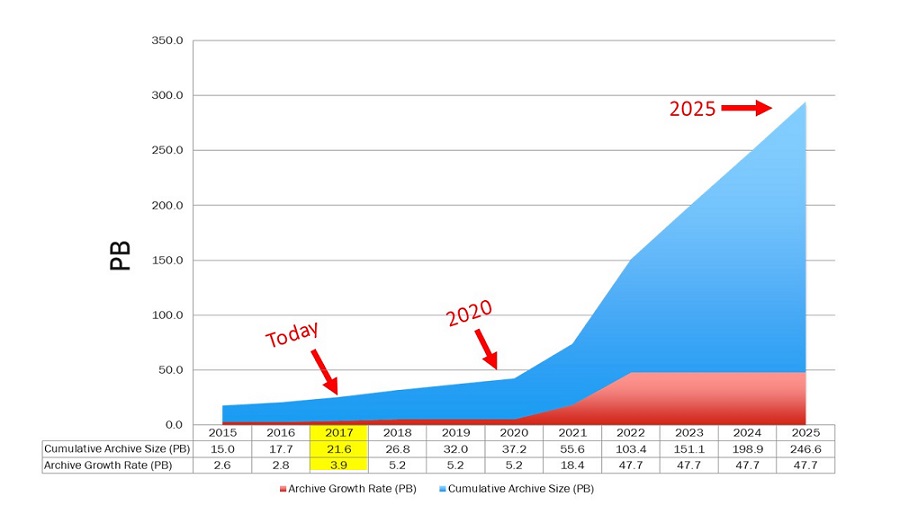
Data volumes:
~5-20 TB/week at steady-state
=> ~1-2 PB to store at steady-state
~10%-20% of that needs to be routinely accessed
Challenges of data-intensive neuroscience
Computational resources
Statistical methods and algorithms
Integration of different data types
Integration of theory and experiments
Patterns of collaboration
Reproducibility and extensibility
What are the requirements?
Bring the compute to the data
Scalable computing
Provide useful tools and interfaces
Facilitate interoperability (between datasets, between software libraries)
Control access
Package into a reproducible unit
Inspiration from other fields of science

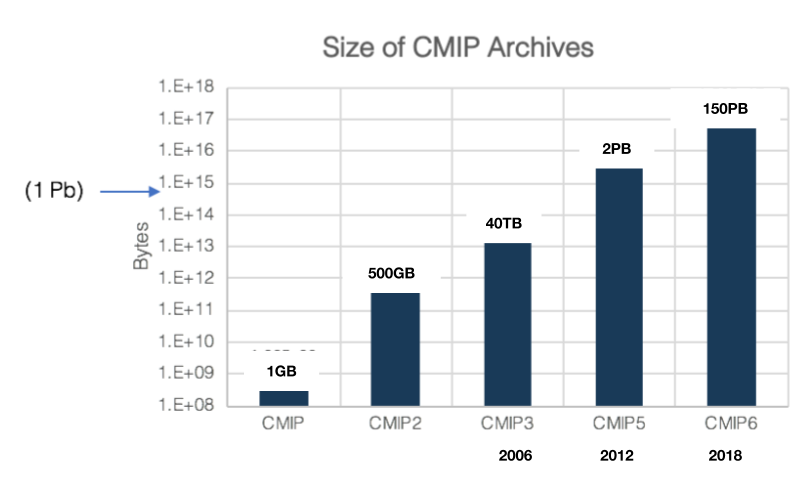
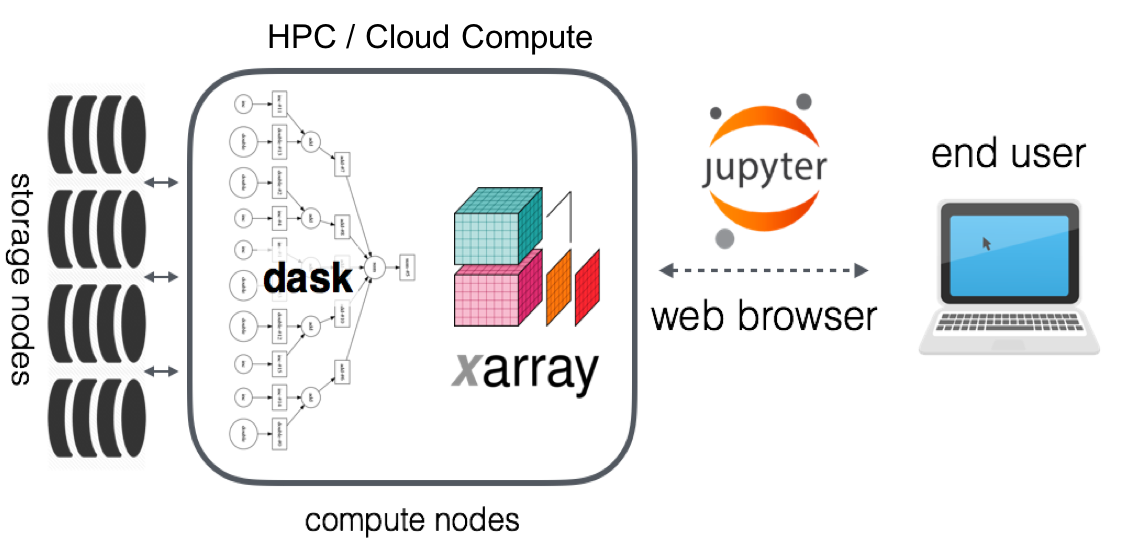
Big data in the geosciences
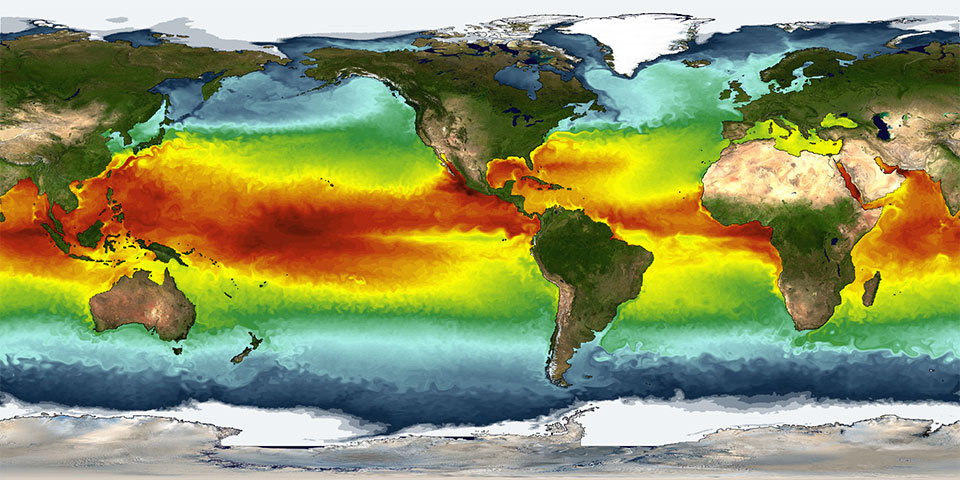
Big data in the geosciences
A community platform for Big Data Geoscience
Open-source scientific computing tools and large open datasets on high-performance computing platforms served through the web-browser
http://pangeo.io
PanNeuro
Cloud computing
Open source scientific computing in Python
Interactive computing with Jupyter
A selection of standardized datasets generated by the collaboration
Why use the cloud?
Data and compute are colocalized
Minimal data transfers
Access control
Consistent and centrally managed
Portable and reproducible
Scientific computing in Python
Python: an ecosystem for scientific computing
Free and open source
High-level interpreted language
Very wide adoption
Both in academic research and in industry
Interactive computing through the web browser
The Jupyter notebook
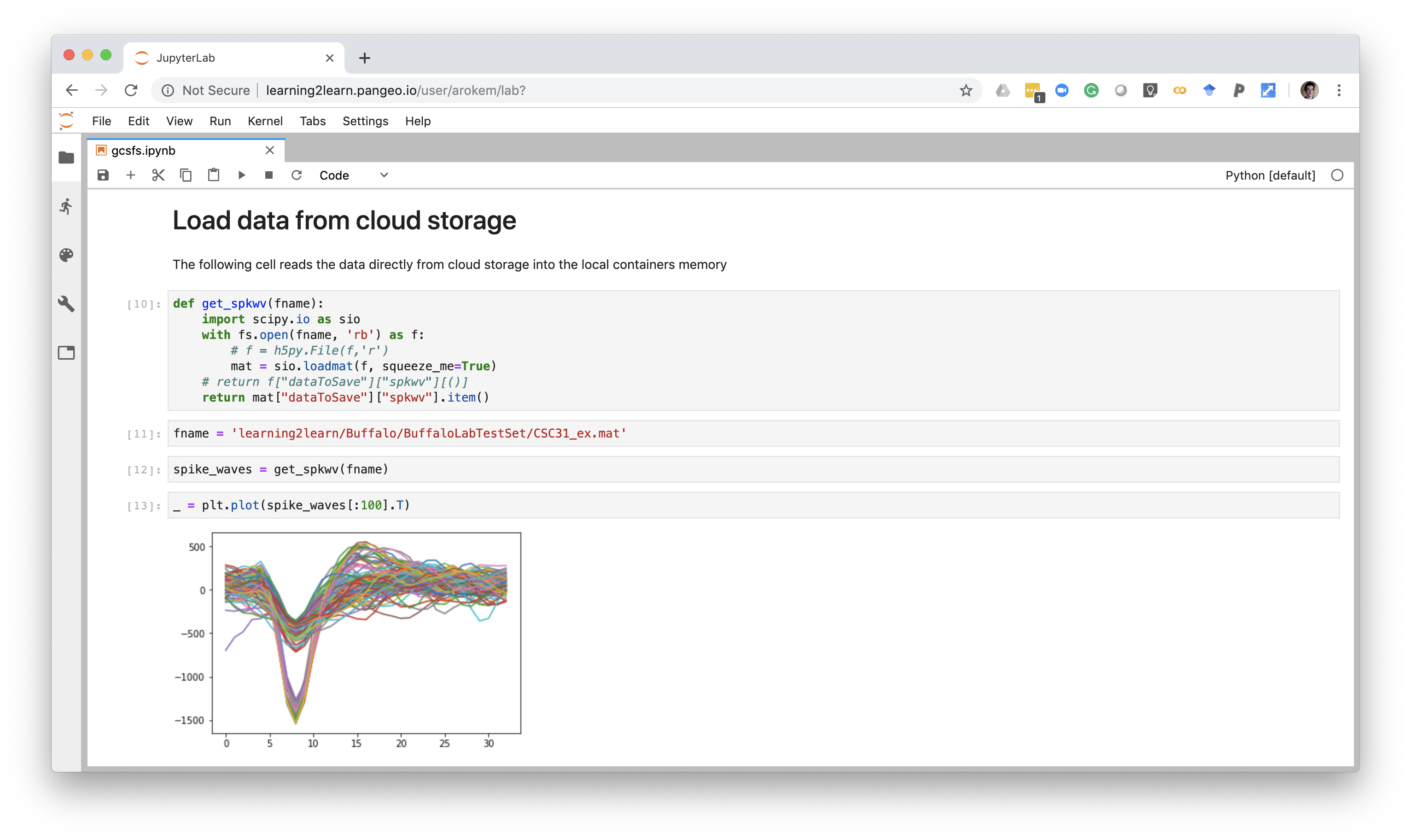
Standardized data formats
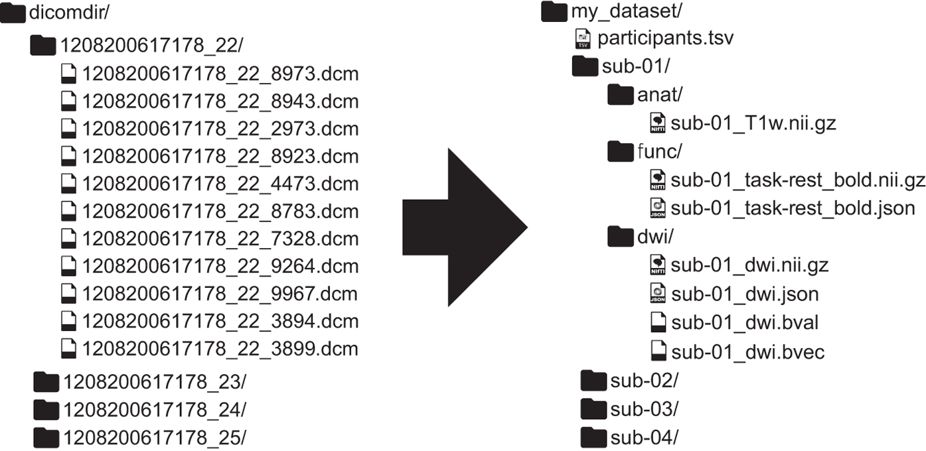
BIDS: a scientist-friendly data standard
Demo

Demo video
Binder demo
Try it out yourself!
http://learning-2-learn.github.io/panneuro_binder_demoBarriers to adoption
Concerns about cost
Reluctance to share data
New skills required
The tools are rapidly evolving
Data formats and data standardization
Conclusions
Leveraging existing investments in Pangeo
Open source software and cyberinfrastructure
Interdisciplinary collaboration
Accessible scalable computing in the browser
The team









We are hiring!

Thanks!


Pangeo is also supported by

Contact information